Distributed Hyperparameter Optimization
Scale up your model tuning with Optuna and Dask

Introduction#
Finding the best hyperparameters for machine learning models can be computationally intensive. This example shows how to use Optuna with Dask to distribute hyperparameter optimization across a cluster, making the search process much faster and more efficient through coordinated parallel execution.
The example performs 500 trials of hyperparameter optimization for an XGBoost classifier in parallel, with results coordinated across the cluster for better convergence. Using Coiled's distributed computing capabilities, what might take hours sequentially can be completed in minutes. You'll need the following packages:
pip install optuna optuna-integration xgboost scikit-learn
pip install optuna optuna-integration xgboost scikit-learn
Full code#
Run this code to start optimizing hyperparameters in parallel. If you're new to Coiled, this will run for free on our account.
import coiled
import optuna
from optuna.integration import DaskStorage
from sklearn.model_selection import cross_val_score, KFold
import xgboost as xgb
import numpy as np
# Create cluster
cluster = coiled.Cluster(
n_workers=50,
worker_cpu=2, # Request 2 CPU cores per worker
worker_memory="8 GiB", # Request 8GB RAM per worker
)
client = cluster.get_client()
# Create synthetic dataset for demonstration
X = np.random.randn(1000, 20)
y = np.random.randint(0, 2, 1000)
# Set up Dask-enabled Optuna storage
backend_storage = optuna.storages.InMemoryStorage()
dask_storage = DaskStorage(storage=backend_storage)
# Create Optuna study with Dask integration
study = optuna.create_study(
direction="maximize",
storage=dask_storage, # Enable distributed coordination
sampler=optuna.samplers.RandomSampler(),
)
def objective(trial):
params = {
"lambda": trial.suggest_float("lambda", 1e-8, 100.0, log=True),
"alpha": trial.suggest_float("alpha", 1e-8, 100.0, log=True),
"colsample_bytree": trial.suggest_float("colsample_bytree", 0.2, 1.0),
"max_depth": trial.suggest_int("max_depth", 2, 10, step=1),
"min_child_weight": trial.suggest_float("min_child_weight", 1e-8, 100, log=True),
"learning_rate": trial.suggest_float("learning_rate", 1e-8, 1.0, log=True),
"gamma": trial.suggest_float("gamma", 1e-8, 1.0, log=True),
"grow_policy": "depthwise",
"eval_metric": "logloss",
}
clf = xgb.XGBClassifier(**params)
fold = KFold(n_splits=5, shuffle=True, random_state=0)
score = cross_val_score(clf, X, y, cv=fold, scoring="neg_log_loss")
return score.mean()
# Run optimization trials in parallel
futures = [client.submit(study.optimize, objective, n_trials=1, pure=False)
for _ in range(500)]
_ = client.gather(futures)
# Get best parameters
print("Best parameters:", study.best_params)
print("Best score:", study.best_value)
# Clean up
cluster.shutdown()
import coiled
import optuna
from optuna.integration import DaskStorage
from sklearn.model_selection import cross_val_score, KFold
import xgboost as xgb
import numpy as np
# Create cluster
cluster = coiled.Cluster(
n_workers=50,
worker_cpu=2, # Request 2 CPU cores per worker
worker_memory="8 GiB", # Request 8GB RAM per worker
)
client = cluster.get_client()
# Create synthetic dataset for demonstration
X = np.random.randn(1000, 20)
y = np.random.randint(0, 2, 1000)
# Set up Dask-enabled Optuna storage
backend_storage = optuna.storages.InMemoryStorage()
dask_storage = DaskStorage(storage=backend_storage)
# Create Optuna study with Dask integration
study = optuna.create_study(
direction="maximize",
storage=dask_storage, # Enable distributed coordination
sampler=optuna.samplers.RandomSampler(),
)
def objective(trial):
params = {
"lambda": trial.suggest_float("lambda", 1e-8, 100.0, log=True),
"alpha": trial.suggest_float("alpha", 1e-8, 100.0, log=True),
"colsample_bytree": trial.suggest_float("colsample_bytree", 0.2, 1.0),
"max_depth": trial.suggest_int("max_depth", 2, 10, step=1),
"min_child_weight": trial.suggest_float("min_child_weight", 1e-8, 100, log=True),
"learning_rate": trial.suggest_float("learning_rate", 1e-8, 1.0, log=True),
"gamma": trial.suggest_float("gamma", 1e-8, 1.0, log=True),
"grow_policy": "depthwise",
"eval_metric": "logloss",
}
clf = xgb.XGBClassifier(**params)
fold = KFold(n_splits=5, shuffle=True, random_state=0)
score = cross_val_score(clf, X, y, cv=fold, scoring="neg_log_loss")
return score.mean()
# Run optimization trials in parallel
futures = [client.submit(study.optimize, objective, n_trials=1, pure=False)
for _ in range(500)]
_ = client.gather(futures)
# Get best parameters
print("Best parameters:", study.best_params)
print("Best score:", study.best_value)
# Clean up
cluster.shutdown()
After running this code, we'll explore how it works and what makes it efficient.
The Problem#
Hyperparameter optimization is a crucial but computationally expensive part of machine learning. For complex models like XGBoost, there can be many hyperparameters to tune, and each combination needs to be evaluated through cross-validation. This creates two challenges:
- The search space is large - we have 7 different hyperparameters to optimize
- Each evaluation requires training multiple models for cross-validation
- Each worker needs sufficient CPU and memory to handle model training
Running this sequentially on a single machine would take hours or even days. By distributing the work across a cluster using Dask we can dramatically speed up the process.
Setting Up the Cluster#
We'll use Coiled to create a cluster with 10 workers, each with 2 CPU cores and 8GB of RAM.
cluster = coiled.Cluster(
n_workers=50, # Run 50 parallel workers
worker_cpu=2, # 2 CPU cores per worker for model training
worker_memory="8 GiB", # 8GB RAM per worker
)
cluster = coiled.Cluster(
n_workers=50, # Run 50 parallel workers
worker_cpu=2, # 2 CPU cores per worker for model training
worker_memory="8 GiB", # 8GB RAM per worker
)
This gives us 100 total CPU cores and 800GB of RAM distributed across our workers, allowing us to efficiently handle parallel model training and evaluation. We could make this much larger if we needed to.
Setting Up the Optimization#
The example uses Optuna with its Dask integration for distributed hyperparameter optimization. The key to efficient distributed execution is the DaskStorage backend:
# Create a shared storage backend and Optuna study
backend_storage = optuna.storages.InMemoryStorage()
dask_storage = DaskStorage(storage=backend_storage)
study = optuna.create_study(
direction="maximize",
storage=dask_storage,
sampler=optuna.samplers.RandomSampler(),
)
# Create a shared storage backend and Optuna study
backend_storage = optuna.storages.InMemoryStorage()
dask_storage = DaskStorage(storage=backend_storage)
study = optuna.create_study(
direction="maximize",
storage=dask_storage,
sampler=optuna.samplers.RandomSampler(),
)
This setup enables:
- Coordinated storage of trial results across the cluster
- Efficient sharing of optimization history between workers
- Better convergence through synchronized parameter sampling
The objective function defines our search space and evaluation metric:
- We optimize 7 key XGBoost parameters including learning rate, regularization terms, and tree parameters
- Each parameter has a defined range and sampling strategy (e.g., log-uniform for learning rate)
- We use 5-fold cross-validation with log loss as our metric
Parallel Execution#
The combination of Dask and Optuna's DaskStorage enables highly efficient parallel optimization:
# Run 500 trials in parallel
futures = [client.submit(
study.optimize, objective, n_trials=1, pure=False)
for _ in range(500)
]
# Run 500 trials in parallel
futures = [client.submit(
study.optimize, objective, n_trials=1, pure=False)
for _ in range(500)
]
Each worker in our cluster can:
- Sample new hyperparameters based on the shared optimization history
- Train and evaluate an XGBoost model
- Report results back to the centralized storage
- Influence future parameter sampling across all workers
This coordinated approach leads to faster convergence compared to independent parallel trials, as each worker benefits from the knowledge gained by others.
Results#
The optimization process completed 500 trials across the cluster, finding optimal hyperparameters for our XGBoost classifier. The best configuration achieved a negative log loss of -0.097, with the following parameters:
print(study.best_params)
print(study.best_params)
{
'lambda': 1.162e-05,
'alpha': 9.831e-05,
'colsample_bytree': 0.219,
'max_depth': 6,
'min_child_weight': 0.816,
'learning_rate': 0.514,
'gamma': 2.756e-05
}
{
'lambda': 1.162e-05,
'alpha': 9.831e-05,
'colsample_bytree': 0.219,
'max_depth': 6,
'min_child_weight': 0.816,
'learning_rate': 0.514,
'gamma': 2.756e-05
}
The optimization history shows steady improvement in model performance across trials:
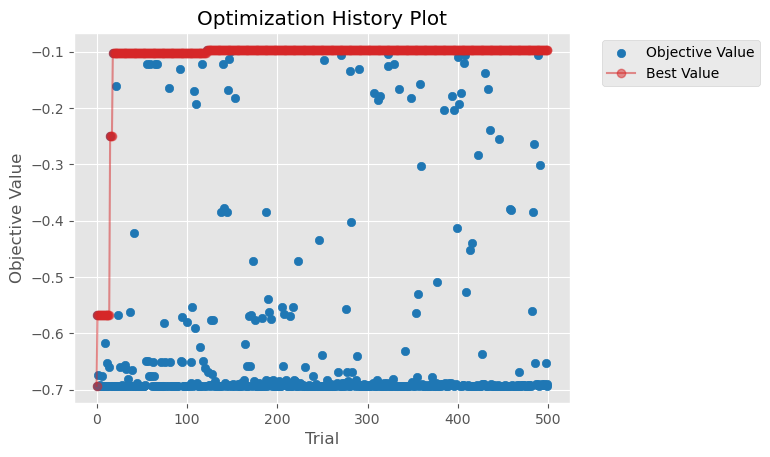
Next Steps#
You can extend this example in several ways:
- Try different hyperparameter ranges or add additional parameters to optimize
- Scale up to more trials by increasing the cluster size
- Apply this pattern to other machine learning models
- Check out our extended XGBoost HPO example for a more detailed implementation
- Explore other machine learning examples with Coiled
Get started
Know Python? Come use the cloud. Your first $25 of usage per month is on us.
$ pip install coiled
$ coiled quickstart
Grant cloud access? (Y/n): Y
... Configuring ...
You're ready to go. 🎉$ pip install coiled
$ coiled quickstart
Grant cloud access? (Y/n): Y
... Configuring ...
You're ready to go. 🎉